
Hi, I'm Farhan Siddiqui
Senior AI Engineer
Transforming businesses with cutting-edge AI solutions. Specialized in Agentic AI, Generative AI, and Full-Stack AI Development. 100+ projects delivered across global organizations.
AI in Healthcare with MedGemma
Healthcare is undergoing a revolutionary transformation with the advent of sophisticated AI models designed specifically for medical applications. Google Research has announced new multimodal models in the MedGemma collection, their most capable open models for health AI development, marking a significant leap forward in healthcare AI capabilities.
Interactive Healthcare AI Demonstrations
Before diving into the technical details, you can experience MedGemma's capabilities firsthand through these interactive demonstrations:
- Appoint Ready Demo: A simulation showing how MedGemma can be built into an application to streamline pre-visit information gathering ahead of a patient appointment
- HAI-DEF Concept Apps Collection: A collection of concept apps built around HAI-DEF open models including pathology image access, radiology report explainers, and pre-visit intake demos
For more information please visit Google Research
The Healthcare AI Revolution
Healthcare is increasingly embracing AI to improve workflow management, patient communication, and diagnostic and treatment support. The introduction of MedGemma represents a paradigm shift in how we approach healthcare AI development, offering developers unprecedented access to powerful, specialized medical AI models.
Health AI Developer Foundations (HAI-DEF)
HAI-DEF is a collection of lightweight open models designed to offer developers robust starting points for their own health research and application development. What makes this particularly valuable is that because HAI-DEF models are open, developers retain full control over privacy, infrastructure and modifications to the models.
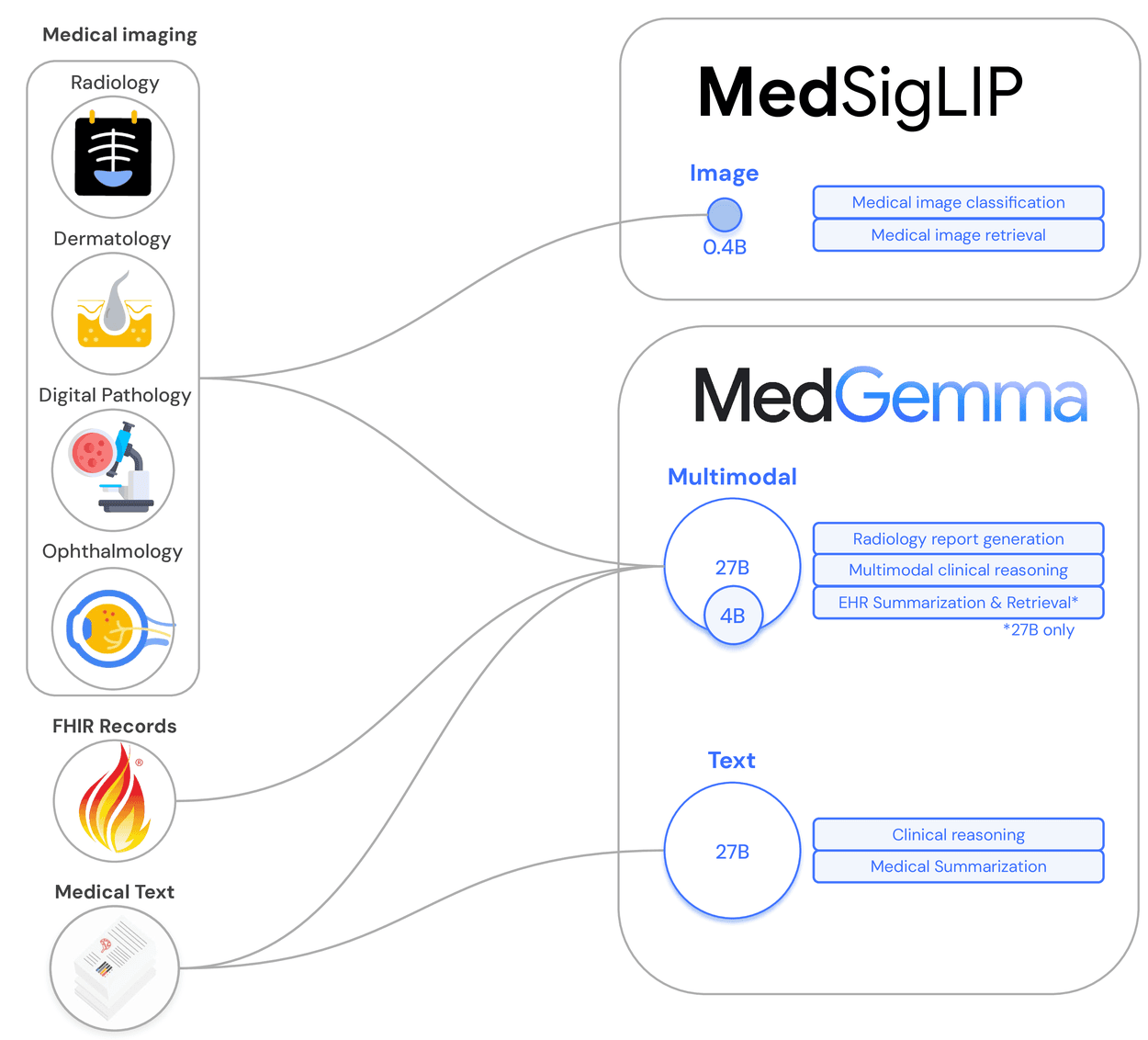
MedGemma: The Multimodal Healthcare AI Powerhouse
Model Variants and Capabilities
The MedGemma collection includes several specialized variants:
MedGemma 4B Multimodal:
- Scores 64.4% on MedQA, ranking it among the best very small (<8B) open models
- In an unblinded study, 81% of MedGemma 4B–generated chest X-ray reports were judged by a US board certified radiologist to be of sufficient accuracy to result in similar patient management compared to the original radiologist reports
- Can be adapted to run on mobile hardware
MedGemma 27B Models:
- Based on internal and published evaluations, the MedGemma 27B models are among the best performing small open models (<50B) on the MedQA medical knowledge and reasoning benchmark
- The text variant scores 87.7%, which is within 3 points of DeepSeek R1, a leading open model, but at approximately one tenth the inference cost
- Competitive with larger models across a variety of benchmarks, including retrieval and interpretation of electronic health record data
Architecture and Training
from transformers import AutoModelForCausalLM, AutoTokenizer import torch class MedGemmaProcessor: def __init__(self, model_size="4b"): self.model_name = f"google/medgemma-{model_size}" self.tokenizer = AutoTokenizer.from_pretrained(self.model_name) self.model = AutoModelForCausalLM.from_pretrained( self.model_name, torch_dtype=torch.float16, device_map="auto" ) async def analyze_medical_image(self, image_path, question): # Process medical image with multimodal capabilities inputs = self.tokenizer( f"Analyze this medical image: {question}", return_tensors="pt" ) with torch.no_grad(): outputs = self.model.generate( inputs.input_ids, max_length=512, temperature=0.7, do_sample=True ) response = self.tokenizer.decode(outputs[0], skip_special_tokens=True) return response async def generate_medical_report(self, patient_data): # Generate comprehensive medical reports prompt = f"Generate a medical report based on: {patient_data}" inputs = self.tokenizer(prompt, return_tensors="pt") with torch.no_grad(): outputs = self.model.generate( inputs.input_ids, max_length=1024, temperature=0.3, do_sample=True ) report = self.tokenizer.decode(outputs[0], skip_special_tokens=True) return report
The models were developed by training a medically optimized image encoder, followed by training the corresponding 4B and 27B versions of the Gemma 3 model on medical data. Importantly, care was taken to retain the general (non-medical) capabilities of Gemma throughout this process, allowing MedGemma to perform well on tasks that mix medical and non-medical information.
MedSigLIP: Specialized Medical Image Understanding
MedSigLIP is a lightweight image encoder of only 400M parameters that uses the Sigmoid loss for Language Image Pre-training (SigLIP) architecture. This specialized model addresses critical healthcare imaging needs:
Key Capabilities
- Traditional image classification: Build performant models to classify medical images
- Zero-shot image classification: Classify images without specific training examples
- Semantic image retrieval: Find visually or semantically similar images from large medical image databases
MedSigLIP was adapted from SigLIP via tuning with diverse medical imaging data, including chest X-rays, histopathology patches, dermatology images, and fundus images.
from transformers import AutoModel, AutoProcessor import torch class MedSigLIPProcessor: def __init__(self): self.model = AutoModel.from_pretrained("google/medsiglip-400m") self.processor = AutoProcessor.from_pretrained("google/medsiglip-400m") async def classify_medical_image(self, image, text_labels): # Zero-shot classification of medical images inputs = self.processor( text=text_labels, images=image, return_tensors="pt", padding=True ) with torch.no_grad(): outputs = self.model(**inputs) logits = outputs.logits_per_image probs = torch.softmax(logits, dim=-1) return probs.cpu().numpy() async def retrieve_similar_images(self, query_image, image_database): # Semantic image retrieval for medical databases query_embedding = self.get_image_embedding(query_image) similarities = [] for db_image in image_database: db_embedding = self.get_image_embedding(db_image) similarity = torch.cosine_similarity( query_embedding, db_embedding, dim=0 ) similarities.append(similarity.item()) return similarities
Real-World Healthcare Applications
Medical Imaging and Diagnostics
After fine-tuning, MedGemma 4B is able to achieve state-of-the-art performance on chest X-ray report generation, with a RadGraph F1 score of 30.3. This capability enables:
- Automated Radiology Reporting: Generate comprehensive reports from medical images
- Diagnostic Assistance: Provide preliminary analysis to support clinical decisions
- Quality Assurance: Cross-reference human interpretations with AI analysis
Electronic Health Records Processing
The MedGemma 27B models are competitive with larger models across a variety of benchmarks, including retrieval and interpretation of electronic health record data. Applications include:
- Patient history summarization
- Clinical note generation
- Treatment recommendation systems
- Drug interaction analysis
Pre-visit Patient Intake
class PreVisitIntakeSystem: def __init__(self): self.medgemma = MedGemmaProcessor("27b") self.patient_data = {} async def conduct_intake_interview(self, patient_id): # Automated pre-visit information gathering intake_questions = [ "What symptoms are you experiencing?", "How long have you had these symptoms?", "Are you currently taking any medications?", "Do you have any allergies?", "What is your medical history?" ] responses = [] for question in intake_questions: # Use MedGemma to generate contextual follow-up questions follow_up = await self.medgemma.generate_followup_question( question, responses ) # Process patient response patient_response = await self.get_patient_response(follow_up) responses.append(patient_response) # Generate preliminary assessment assessment = await self.medgemma.generate_medical_report(responses) return assessment async def prioritize_appointments(self, intake_data): # Use AI to prioritize patient appointments based on urgency urgency_score = await self.medgemma.assess_urgency(intake_data) return urgency_score
Developer Success Stories
Developers at DeepHealth in Massachusetts, USA have been exploring MedSigLIP to improve their chest X-ray triaging and nodule detection. Researchers at Chang Gung Memorial Hospital in Taiwan noted that MedGemma works well with traditional Chinese-language medical literature and can respond well to medical staff questions.
Developers at Tap Health in Gurgaon, India, remarked on MedGemma's superior medical grounding, noting its reliability on tasks that require sensitivity to clinical context, such as summarizing progress notes or suggesting guideline-aligned nudges.
Advantages of Open Healthcare AI Models
Because the MedGemma collection is open, the models can be downloaded, built upon, and fine-tuned to support developers' specific needs. This approach offers several distinct advantages:
Privacy and Control
- Models can be run on proprietary hardware in the developer's preferred environment, including on Google Cloud Platform or locally, which can address privacy concerns or institutional policies
- Full control over patient data processing and storage
- Compliance with healthcare regulations like HIPAA
Customization and Performance
- Models can be fine-tuned and modified to achieve optimal performance on target tasks and datasets
- Specialized training for specific medical domains
- Integration with existing healthcare systems
Stability and Reproducibility
- Because the models are distributed as snapshots, their parameters are frozen and unlike an API, will not change unexpectedly over time
- This stability is particularly crucial for medical applications where consistency and reproducibility are paramount
Implementation Best Practices
Model Selection Guidelines
- MedGemma 4B: Ideal for mobile applications and resource-constrained environments
- MedGemma 27B: Best for complex medical reasoning and comprehensive analysis
- MedSigLIP: Perfect for medical image classification and retrieval tasks
Performance Optimization
class OptimizedMedGemmaDeployment: def __init__(self): self.model_cache = {} self.gpu_memory_manager = GPUMemoryManager() async def load_model_efficiently(self, model_type, task_type): # Efficient model loading based on task requirements if task_type == "image_classification": if "medsiglip" not in self.model_cache: self.model_cache["medsiglip"] = MedSigLIPProcessor() return self.model_cache["medsiglip"] elif task_type == "text_generation": model_size = "4b" if self.gpu_memory_manager.is_constrained() else "27b" cache_key = f"medgemma_{model_size}" if cache_key not in self.model_cache: self.model_cache[cache_key] = MedGemmaProcessor(model_size) return self.model_cache[cache_key] async def batch_process_medical_data(self, data_batch): # Efficient batch processing for medical data results = [] for data_item in data_batch: model = await self.load_model_efficiently( data_item.model_type, data_item.task_type ) result = await model.process(data_item) results.append(result) return results
Clinical Integration Considerations
-
Validation and Testing: MedGemma and MedSigLIP are intended to be used as a starting point that enables efficient development of downstream healthcare applications involving medical text and images
-
Clinical Oversight: The outputs generated by these models are not intended to directly inform clinical diagnosis, patient management decisions, treatment recommendations, or any other direct clinical practice applications
-
Verification Requirements: All model outputs should be considered preliminary and require independent verification, clinical correlation, and further investigation
Future Implications and Trends
Enhanced Multimodal Capabilities
The integration of text and image processing in MedGemma opens new possibilities for:
- Comprehensive Patient Assessment: Combining lab results, imaging, and clinical notes
- Longitudinal Care Tracking: Monitoring patient progress over time
- Predictive Healthcare: Early warning systems for potential health issues
Democratization of Healthcare AI
With MedGemma 4B and MedSigLIP able to run on mobile hardware, we're moving toward:
- Point-of-care diagnostics in remote areas
- Personal health monitoring applications
- Real-time clinical decision support
Global Healthcare Impact
The multilingual capabilities of MedGemma, as demonstrated by its effectiveness with traditional Chinese-language medical literature, suggest potential for:
- Cross-cultural medical knowledge sharing
- Standardized healthcare quality worldwide
- Reduced healthcare disparities in underserved regions
Getting Started with MedGemma
Development Resources
Detailed notebooks are provided on GitHub for MedGemma and MedSigLIP that demonstrate how to create instances for both inference and fine-tuning on Hugging Face. When developers are ready to scale, MedGemma and MedSigLIP can be seamlessly deployed in Vertex AI as dedicated endpoints.
Community and Support
The HAI-DEF forum is available for questions or feedback, providing a collaborative environment for healthcare AI developers to share experiences and best practices.
The introduction of MedGemma and MedSigLIP represents a watershed moment in healthcare AI development. By providing open, capable, and specialized models, Google Research has democratized access to sophisticated healthcare AI tools, enabling developers worldwide to create innovative solutions that can transform patient care, clinical workflows, and medical research. The future of healthcare AI is not just about powerful models—it's about making those models accessible, adaptable, and actionable for real-world healthcare challenges.
Ready to Transform Your Business with AI?
Let's discuss how we can implement these AI solutions for your organization.
Get Started